This is the Linux app named MANTI.pl / muda.pl whose latest release can be downloaded as muda.pl-v3.2.zip. It can be run online in the free hosting provider OnWorks for workstations.
Download and run online this app named MANTI.pl / muda.pl with OnWorks for free.
Follow these instructions in order to run this app:
- 1. Downloaded this application in your PC.
- 2. Enter in our file manager https://www.onworks.net/myfiles.php?username=XXXXX with the username that you want.
- 3. Upload this application in such filemanager.
- 4. Start the OnWorks Linux online or Windows online emulator or MACOS online emulator from this website.
- 5. From the OnWorks Linux OS you have just started, goto our file manager https://www.onworks.net/myfiles.php?username=XXXXX with the username that you want.
- 6. Download the application, install it and run it.
SCREENSHOTS
Ad
MANTI.pl / muda.pl
DESCRIPTION
-------- ATTENTION START: RENAMING
muda.pl was renamed to MANTI.pl with v3.7, project development can be tracked on the MANTI project page on sourceforge.net. Old versions remain here for archival purposes.
-------- ATTENTION END
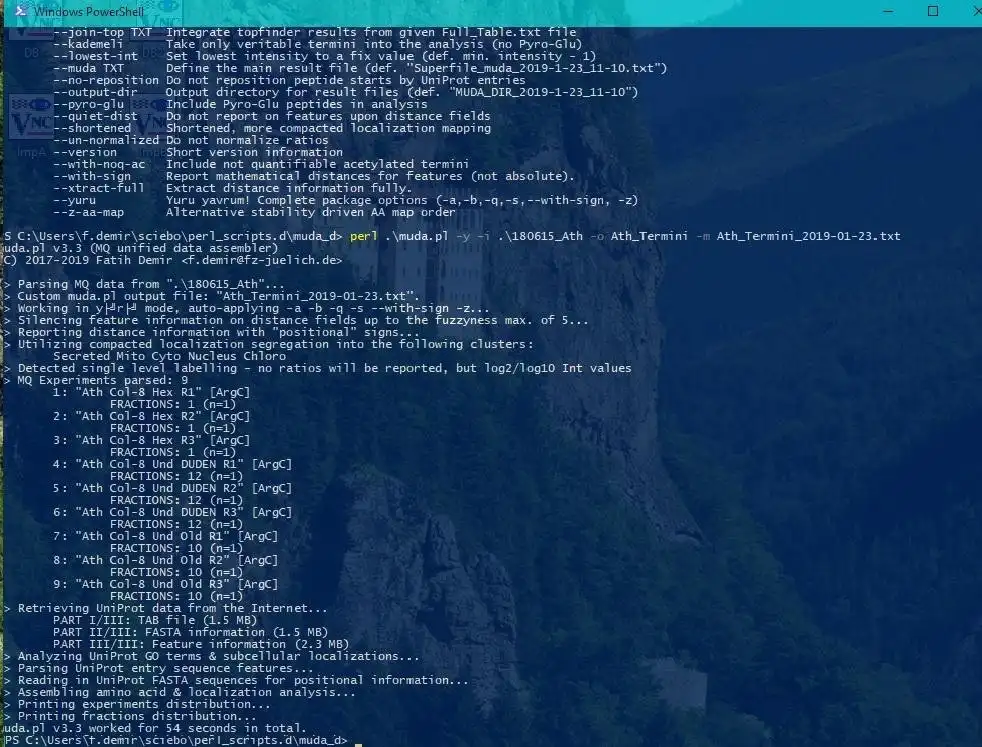
muda.pl is an evaluation script (written in Perl) without great dependencies.
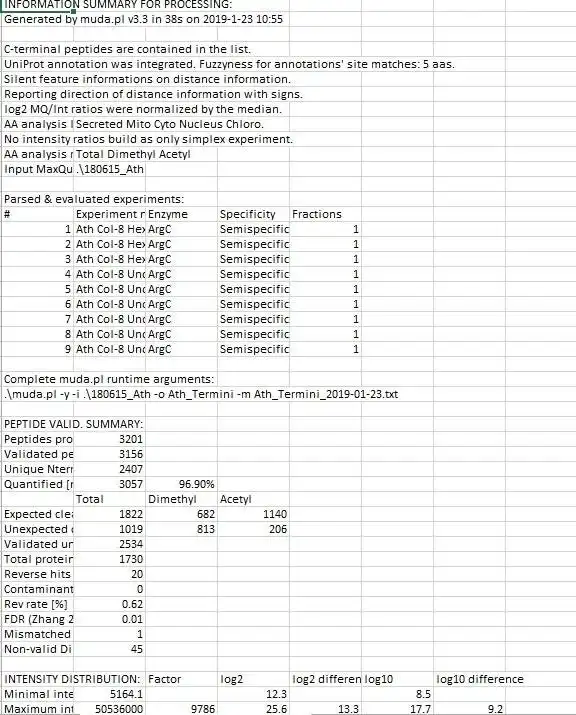
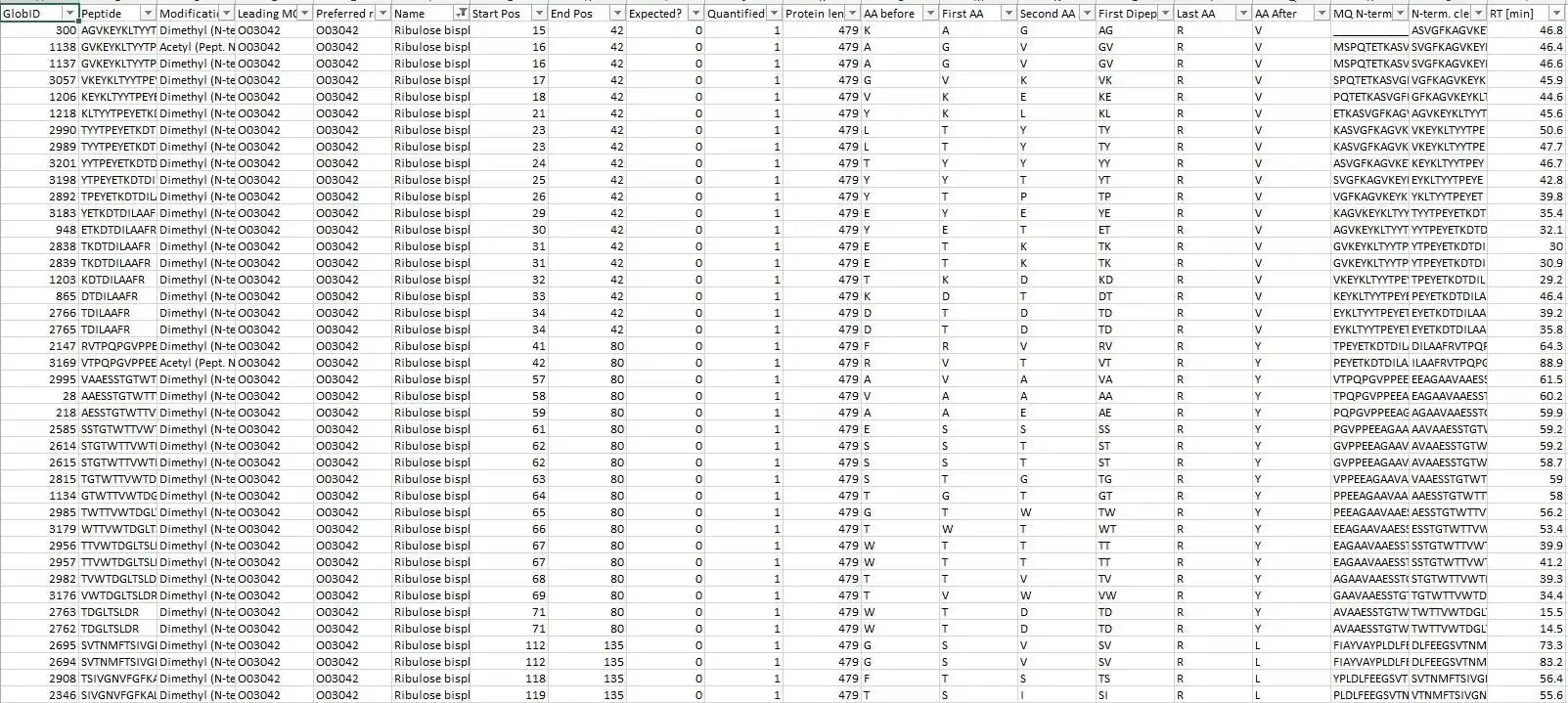
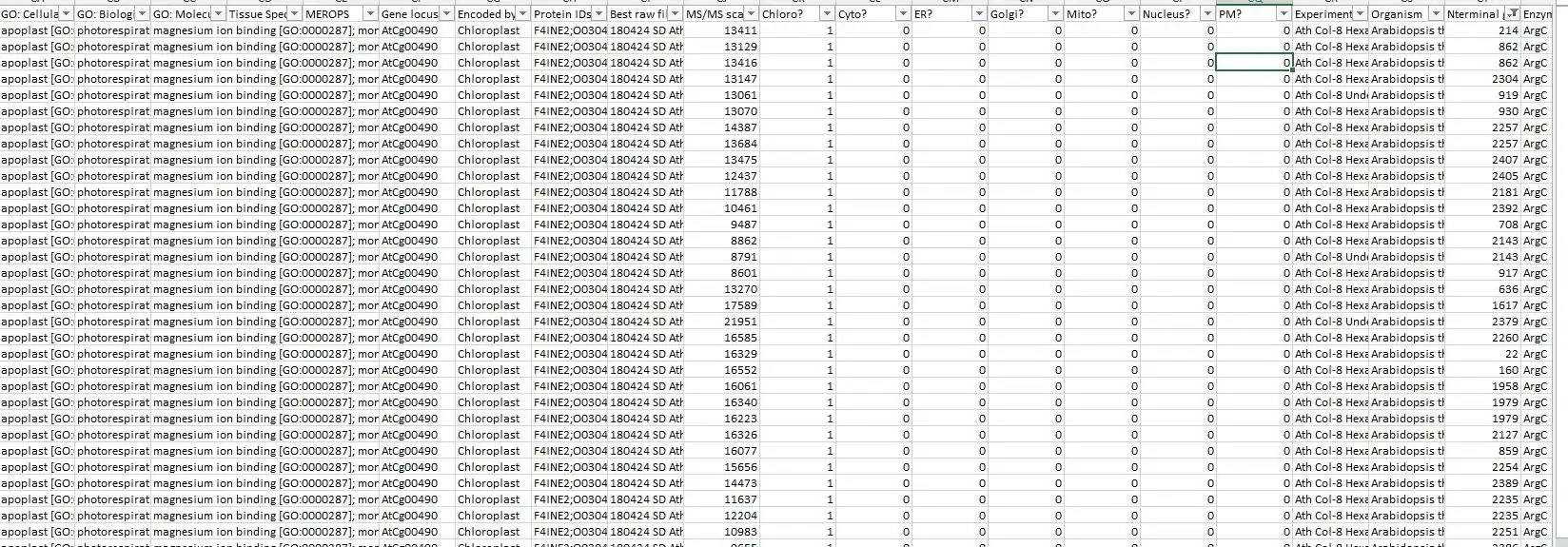
It congregates information from 4 different MaxQuant output files into a master file suitable explicitly for protein neo-termini analyses. The central anchor for the data congregation is the modificationSpecificPeptides.txt file - additional data is inferred from different other source files from the MaxQuant txt folder but the starting point for the data assembly is solely the modificationSpecificPeptides.txt file. Maybe also useful for normal proteomics purposes but this script is heavily optimized for protein neo-termini identification and validation.
For a more thorough explanation of script parameters and evaluation strategy, please consult the extensive manual PDF.
Features
- MaxQuant data evaluation
- Protein-Termini identification & validation
- UniProt data annotation & assembly
- Re-mapping of identified Termini to Isoforms
- Limma, LOCALIZER & TopFINDer integration
- Well documented
Audience
Science/Research
User interface
Command-line
Programming Language
Perl
Categories
This is an application that can also be fetched from https://sourceforge.net/projects/muda/. It has been hosted in OnWorks in order to be run online in an easiest way from one of our free Operative Systems.