This is the Linux app named PhyloTrack to run in Linux online whose latest release can be downloaded as PhyTB.v2.1.zip. It can be run online in the free hosting provider OnWorks for workstations.
Download and run online this app named PhyloTrack to run in Linux online with OnWorks for free.
Follow these instructions in order to run this app:
- 1. Downloaded this application in your PC.
- 2. Enter in our file manager https://www.onworks.net/myfiles.php?username=XXXXX with the username that you want.
- 3. Upload this application in such filemanager.
- 4. Start the OnWorks Linux online or Windows online emulator or MACOS online emulator from this website.
- 5. From the OnWorks Linux OS you have just started, goto our file manager https://www.onworks.net/myfiles.php?username=XXXXX with the username that you want.
- 6. Download the application, install it and run it.
SCREENSHOTS
Ad
PhyloTrack to run in Linux online
DESCRIPTION
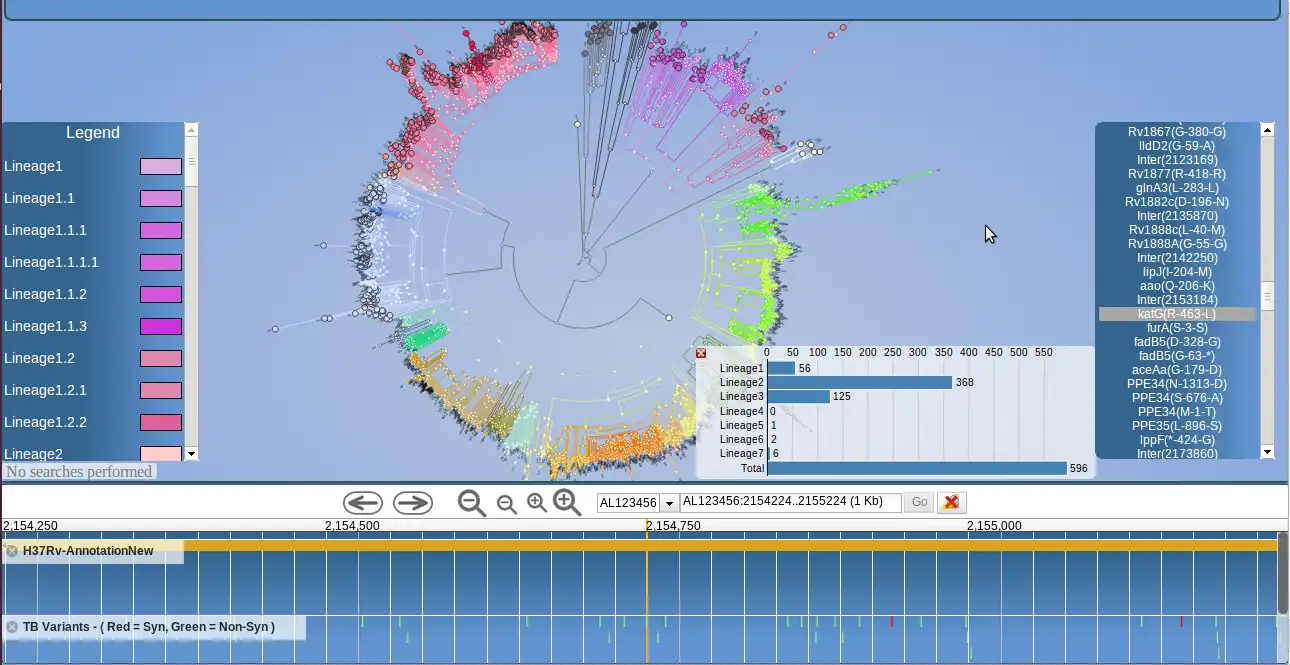
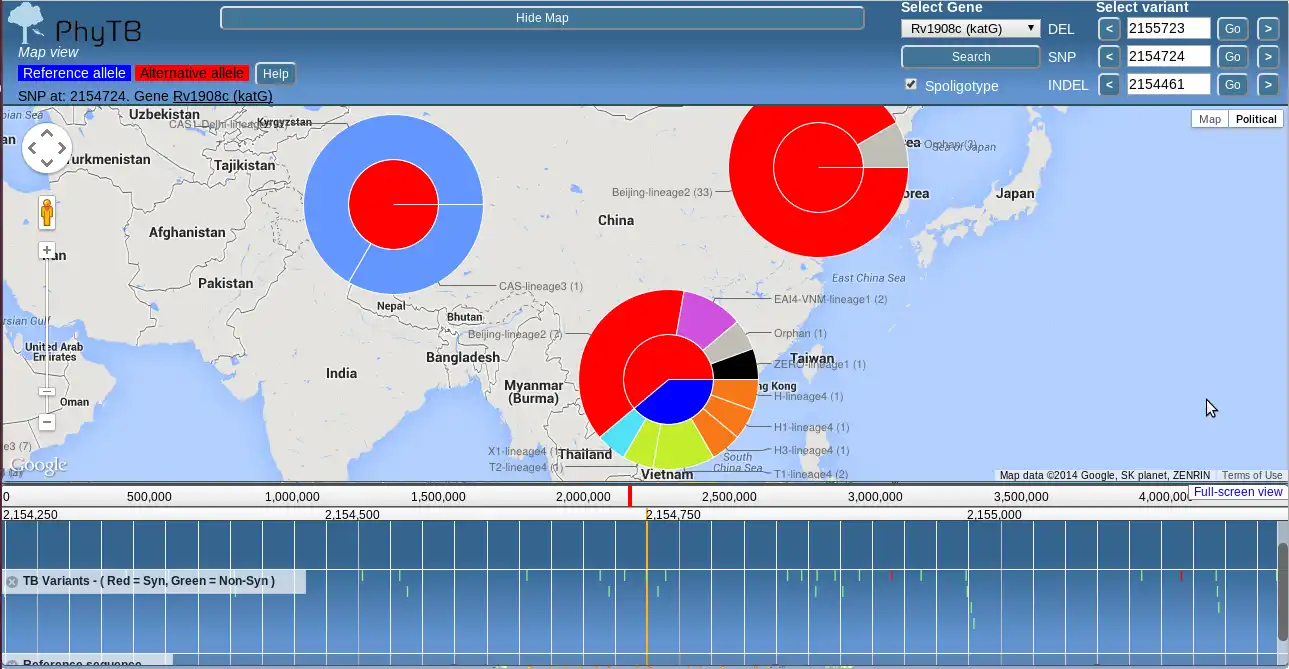
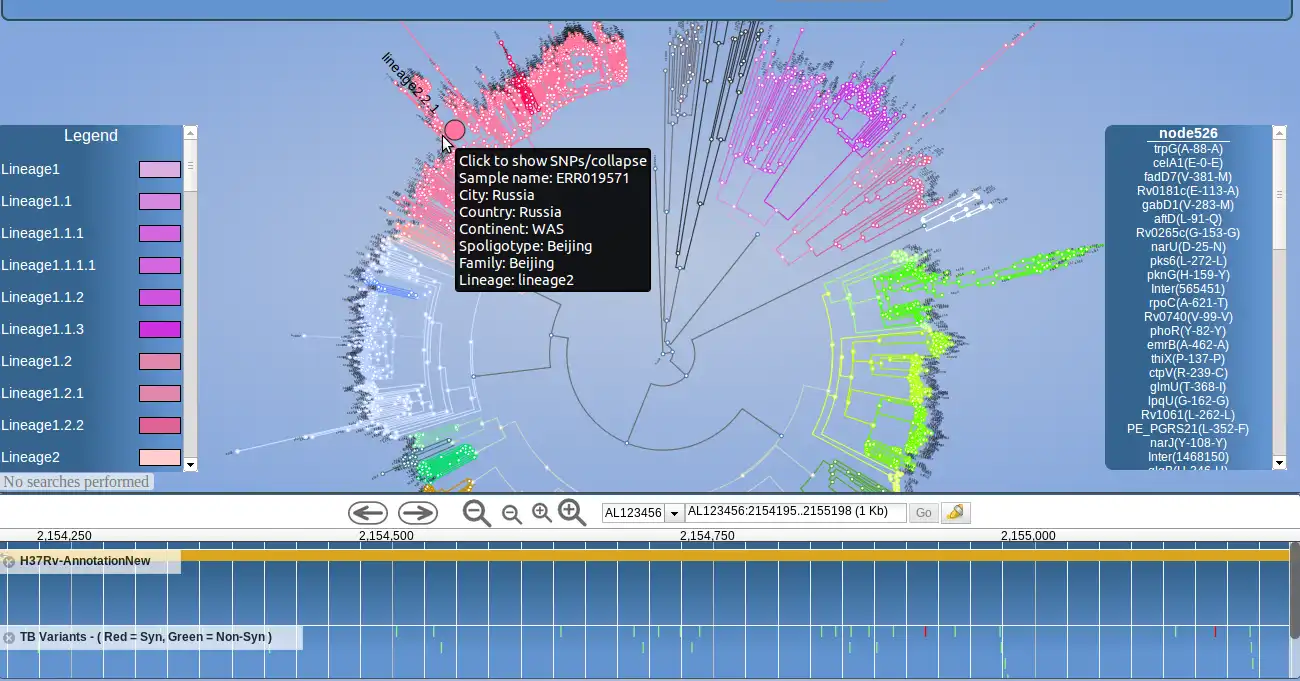
PhyloTrack is a JavaScript--based software tool that integrates the D3.js library for data visualization with the JBrowse tool for genome browser representation. It requires a phylogenetic tree of the common Newick data format as input, as well as three meta data files for samples, clade-defining nodes and clade color definitions - all in tab delimited format. Functionality within PhyloTrack shows the informative markers at each node in the phylogenetic tree, therefore highlighting clade-defining polymorphism. This functionality has been implemented using the tabix tool on the server side, providing simple and rapid access to the information at each tree node, including informative SNPs stored in VCF-similar files. These informative variants have been established by comparing allele frequencies between strain-types using ancestral node comparisons and FST measures of population differentiation.Features
- Phylogeny representation of samples
- Genome browser
- Map distribution of alleles and spoligotypes
- Sample analysis and positioning in the tree
Audience
Science/Research
Programming Language
Python, Perl, PHP, JavaScript
Database Environment
Other file-based DBMS
This is an application that can also be fetched from https://sourceforge.net/projects/phylotrack/. It has been hosted in OnWorks in order to be run online in an easiest way from one of our free Operative Systems.