This is the Linux app named piv_clustering whose latest release can be downloaded as piv_clustering_1.3.tar.gz. It can be run online in the free hosting provider OnWorks for workstations.
Download and run online this app named piv_clustering with OnWorks for free.
Follow these instructions in order to run this app:
- 1. Downloaded this application in your PC.
- 2. Enter in our file manager https://www.onworks.net/myfiles.php?username=XXXXX with the username that you want.
- 3. Upload this application in such filemanager.
- 4. Start the OnWorks Linux online or Windows online emulator or MACOS online emulator from this website.
- 5. From the OnWorks Linux OS you have just started, goto our file manager https://www.onworks.net/myfiles.php?username=XXXXX with the username that you want.
- 6. Download the application, install it and run it.
SCREENSHOTS
Ad
piv_clustering
DESCRIPTION
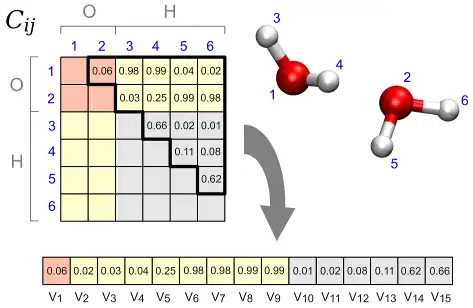
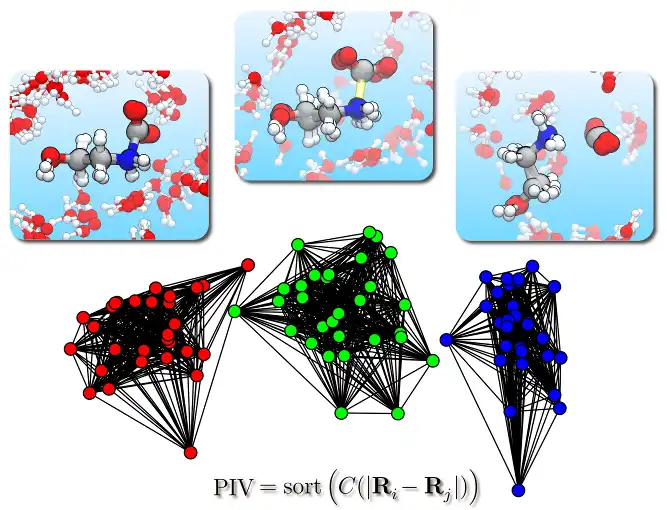
This program allows to perform a structural cluster analysis of atomic trajectories obtained, e.g., from molecular dynamics simulations.
At variance with other approaches, it is possible to analyse also processes in solution, e.g., chemical reactions in liquid water, since the distance metric is based on a Permutation Invariant Vector (PIV) that is symmetric under exchange of identical atoms or molecules, including on the same footing both solute and solvent degrees of freedom. The approach is general and the definition of PIV only requires to specify the range of interatomic distances that is relevant for the process under study. Alternatively, also the SPRINT topological coordinates can be employed, that are particularly suited for nanostructures.
The output is a partitioning of the trajectory into a few structural clusters (i.e., sets of frames), allowing for simpler analysis and visualization.
Please read and cite Gallet & Pietrucci, J. Chem. Phys. 139, 074101 (2013)
Features
- Structural clustering of atomistic trajectories
- Permutation invariance: crystals, amorphous solids, liquids, nanostructures
- All simulation cells are supported (from orthorombic to triclininc)
- Parallel MPI implementation
Audience
Science/Research
Programming Language
Fortran
Categories
This is an application that can also be fetched from https://sourceforge.net/projects/pivclustering/. It has been hosted in OnWorks in order to be run online in an easiest way from one of our free Operative Systems.