This is the Linux app named SPANDx to run in Linux online whose latest release can be downloaded as SPANDx_v3.2.tar.gz. It can be run online in the free hosting provider OnWorks for workstations.
Download and run online this app named SPANDx to run in Linux online with OnWorks for free.
Follow these instructions in order to run this app:
- 1. Downloaded this application in your PC.
- 2. Enter in our file manager https://www.onworks.net/myfiles.php?username=XXXXX with the username that you want.
- 3. Upload this application in such filemanager.
- 4. Start the OnWorks Linux online or Windows online emulator or MACOS online emulator from this website.
- 5. From the OnWorks Linux OS you have just started, goto our file manager https://www.onworks.net/myfiles.php?username=XXXXX with the username that you want.
- 6. Download the application, install it and run it.
SCREENSHOTS
Ad
SPANDx to run in Linux online
DESCRIPTION
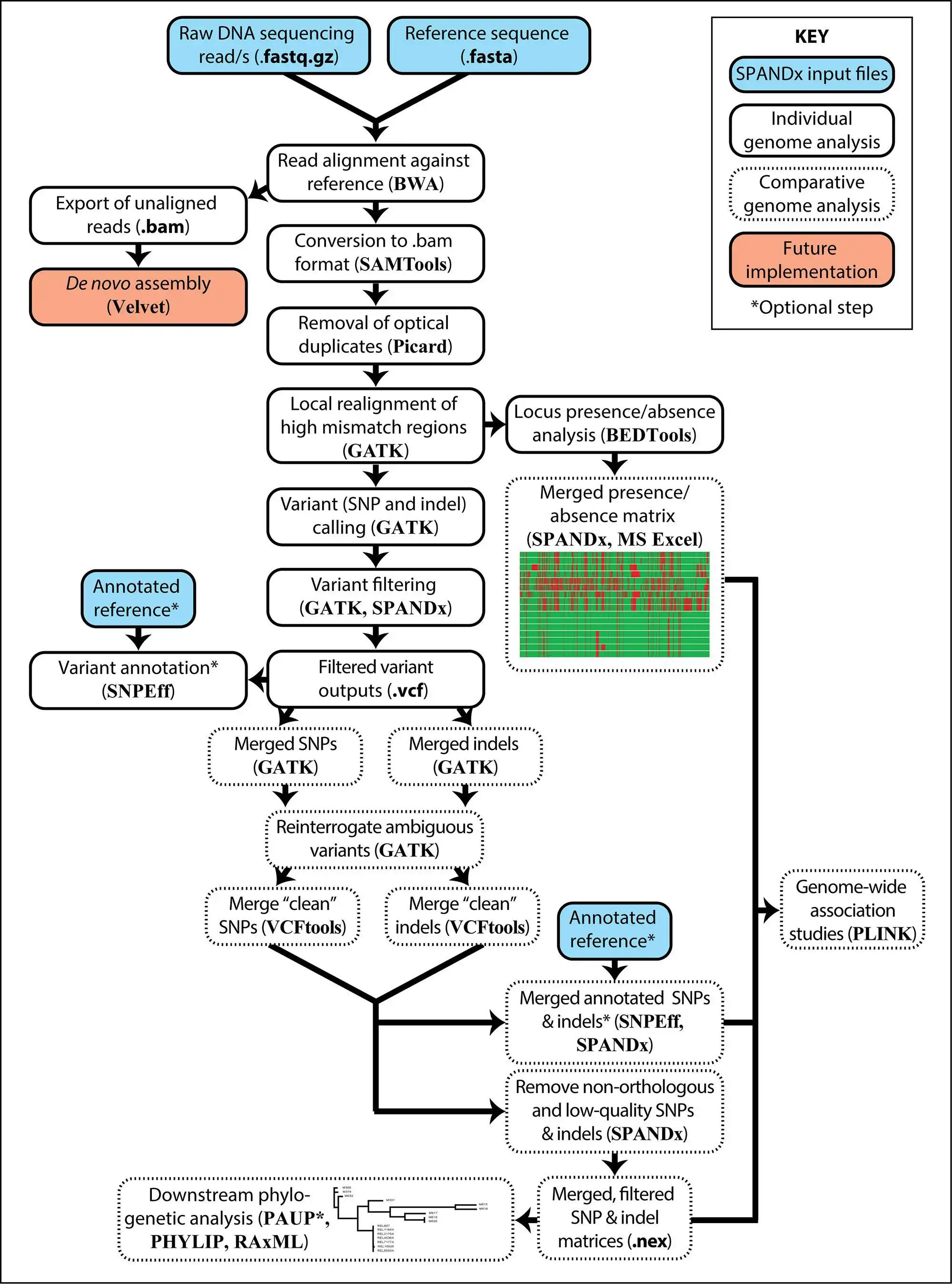
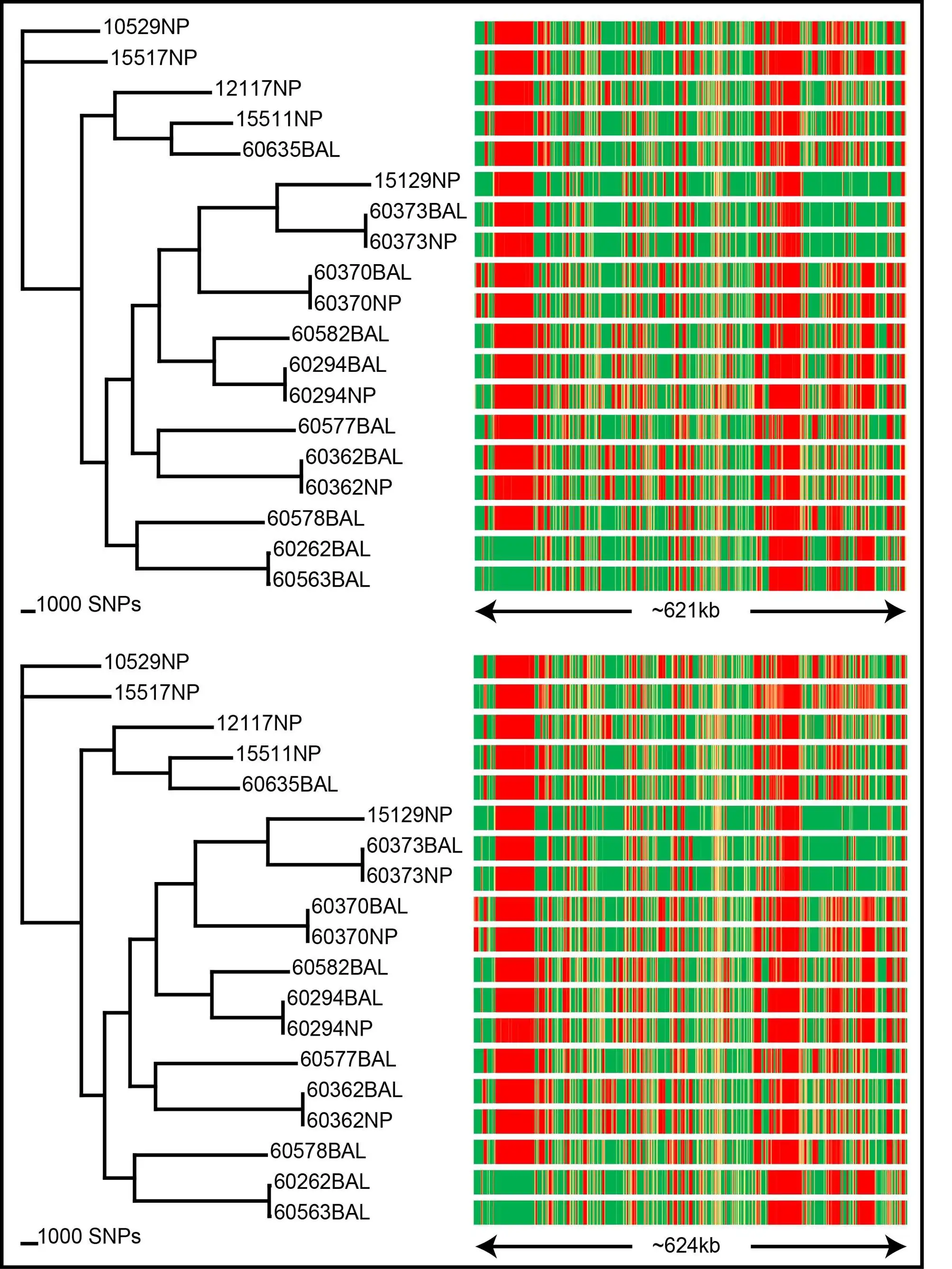
SPANDx is your one-stop tool for identifying SNP and indel variants in haploid genomes using NGS data.SPANDx performs alignment of raw NGS reads against your chosen reference genome or pan-genome, followed by accurate variant calling and annotation, and locus presence/absence determination. SPANDx produces SNP and indel matrices for downstream phylogenetic analyses. Annotated, genome-wide SNPs and indels can also be identified if specified, and are output in human readable format. A presence/absence matrix is also generated to allow you to identify the core/accessory genome content across all your genomes.
The outputs generated by SPANDx can be imported into PLINK for microbial genome-wide association study (mGWAS) analyses.
SPANDx can utilise PBS, SGE or SLURM for resource management. SPANDx can also run directly on the command line if no resource manager is available.
For the most up-to-date version of SPANDx, check us out on GitHub: https://github.com/dsarov/SPANDx
Features
- 30Nov16: SPANDx 3.2 now uses BWA-mem for faster more accurate alignments
- 31May16: SPANDx 3.1.1 now works on pre-v2.3.1 TORQUE/PBS systems.
- 14Feb16: SPANDx 3.1 The -z flag can now be set to include tri- and tetra allelic SNPs in the .nex outputs.
- 26Dec15: SPANDx 3.0 has lots of upgrades including improved samtools compatibility, better PLINK integration for GWAS, and can call up to 9 indels and all four SNPs at a single locus!
- 05Aug15: SPANDx v2.7 now works on SGE, PBS, SLURM, and systems without a resource manager.
Audience
Science/Research
User interface
Command-line
Programming Language
Unix Shell
This is an application that can also be fetched from https://sourceforge.net/projects/spandx/. It has been hosted in OnWorks in order to be run online in an easiest way from one of our free Operative Systems.