This is the Windows app named Dress Up RNA seq to run in Windows online over Linux online whose latest release can be downloaded as DressUp.zip. It can be run online in the free hosting provider OnWorks for workstations.
Download and run online this app named Dress Up RNA seq to run in Windows online over Linux online with OnWorks for free.
Follow these instructions in order to run this app:
- 1. Downloaded this application in your PC.
- 2. Enter in our file manager https://www.onworks.net/myfiles.php?username=XXXXX with the username that you want.
- 3. Upload this application in such filemanager.
- 4. Start any OS OnWorks online emulator from this website, but better Windows online emulator.
- 5. From the OnWorks Windows OS you have just started, goto our file manager https://www.onworks.net/myfiles.php?username=XXXXX with the username that you want.
- 6. Download the application and install it.
- 7. Download Wine from your Linux distributions software repositories. Once installed, you can then double-click the app to run them with Wine. You can also try PlayOnLinux, a fancy interface over Wine that will help you install popular Windows programs and games.
Wine is a way to run Windows software on Linux, but with no Windows required. Wine is an open-source Windows compatibility layer that can run Windows programs directly on any Linux desktop. Essentially, Wine is trying to re-implement enough of Windows from scratch so that it can run all those Windows applications without actually needing Windows.
SCREENSHOTS
Ad
Dress Up RNA seq to run in Windows online over Linux online
DESCRIPTION
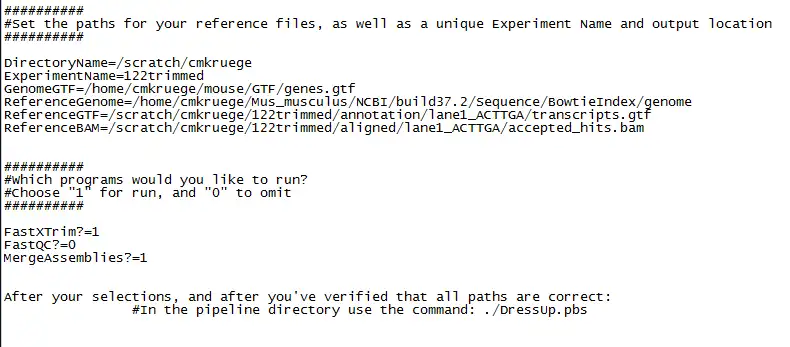
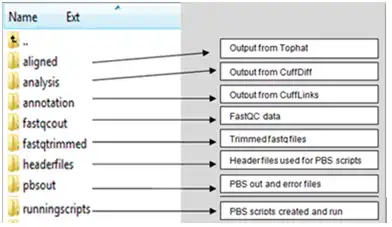
The purpose of DressUp is to create an end-to-end RNA seq pipeline in which all of the steps of analyzing data from an Illumina sequencer is done in one step in an HPC environment. RNA seq programs included are TopHat, CuffLinks, CuffDiff, CuffMerge, FastQC, and trimming using the FastX toolkit.DressUp facilitates RNA seq programs by streamlining various packages into a single script. That script executes the RNA seq programs on a batch cluster system. Upon execution of the script, jobs are submitted to the cluster depending on attributes set up in the configuration file.
Program options can be modified in the main script "DressUp", and PBS options can be modified from the HeaderFiles directory.
Features
- CuffMerge, FastX trimming, and FastQC can be turned on or off from configuration file
- Separate PBS scripts for each component of the pipeline available
- Multiple RNA seq programs performed in one step
- Ability to adjust PBS options for each program
Audience
Science/Research
This is an application that can also be fetched from https://sourceforge.net/projects/dressuprnaseq/. It has been hosted in OnWorks in order to be run online in an easiest way from one of our free Operative Systems.