This is the Windows app named GESPA to run in Windows online over Linux online whose latest release can be downloaded as GESPA.zip. It can be run online in the free hosting provider OnWorks for workstations.
Download and run online this app named GESPA to run in Windows online over Linux online with OnWorks for free.
Follow these instructions in order to run this app:
- 1. Downloaded this application in your PC.
- 2. Enter in our file manager https://www.onworks.net/myfiles.php?username=XXXXX with the username that you want.
- 3. Upload this application in such filemanager.
- 4. Start any OS OnWorks online emulator from this website, but better Windows online emulator.
- 5. From the OnWorks Windows OS you have just started, goto our file manager https://www.onworks.net/myfiles.php?username=XXXXX with the username that you want.
- 6. Download the application and install it.
- 7. Download Wine from your Linux distributions software repositories. Once installed, you can then double-click the app to run them with Wine. You can also try PlayOnLinux, a fancy interface over Wine that will help you install popular Windows programs and games.
Wine is a way to run Windows software on Linux, but with no Windows required. Wine is an open-source Windows compatibility layer that can run Windows programs directly on any Linux desktop. Essentially, Wine is trying to re-implement enough of Windows from scratch so that it can run all those Windows applications without actually needing Windows.
SCREENSHOTS
Ad
GESPA to run in Windows online over Linux online
DESCRIPTION
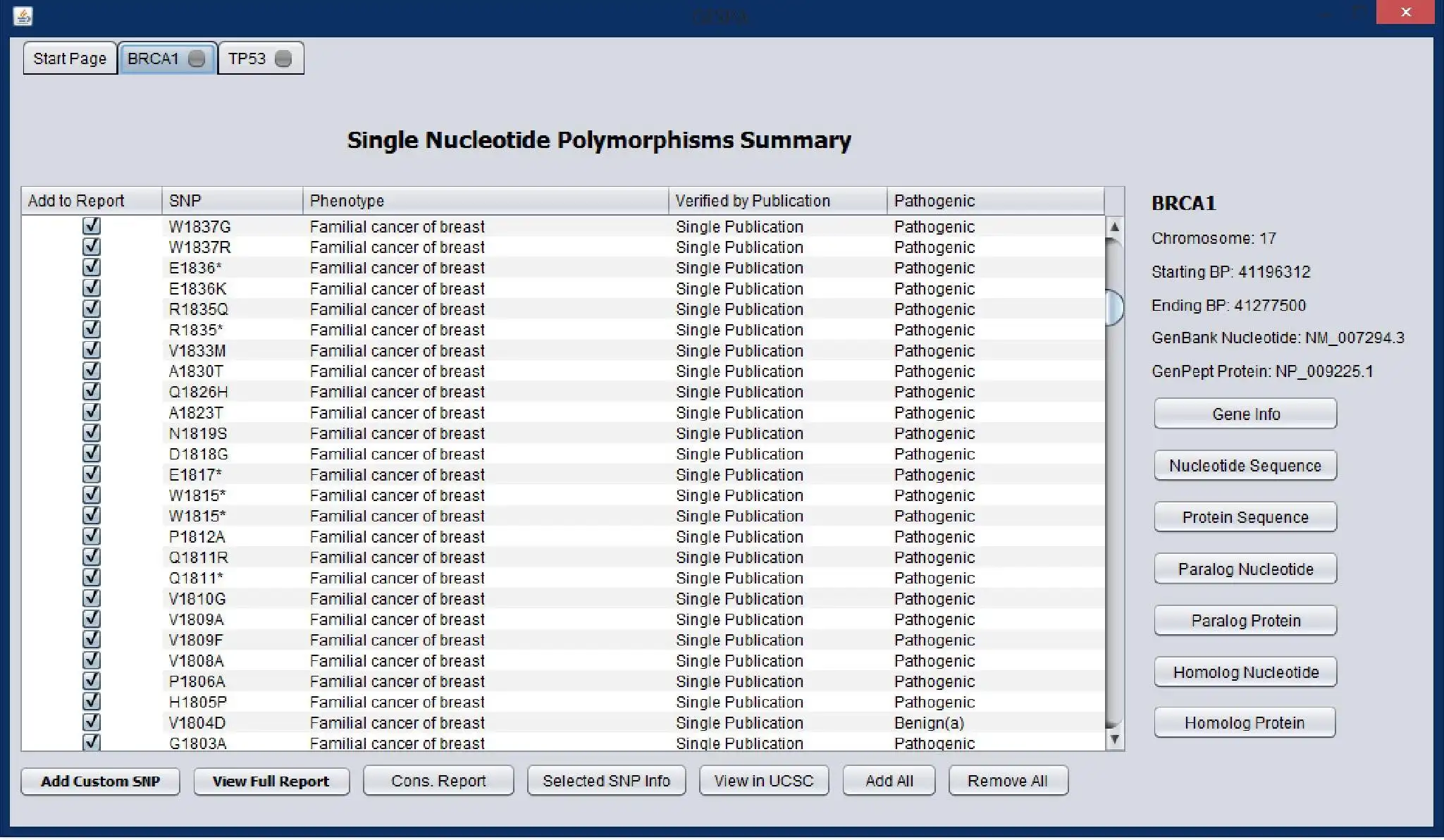
GESPA (GEnomic Single nucleotide Polymorphism Analyzer) is a bioinformatics tool for classifying Nonsynonymous Single Nucleotide Polymorphisms (nsSNPs). GESPA predicts if a nsSNP is pathogenic using reports from literature and various algorithms to assess conservation in orthologous and paralogous protein alignments. Using reports from literature, GESPA is also able to predict the phenotype of a nsSNP with high accuracy. The software can be used clinically to determine if observed nsSNPs are associated with disease. A host of annotations are provided: orthologous and paralogous multiple sequence alignments, UCSC annotations, reports detailing conservation of a nsSNP in alignments, and links to external nsSNP and gene information such as relevant publications. GESPA is connected to a constantly updating SQL server allowing for fast data retrieval. NOTE: REQUIRES Java 1.7.0+. Port 1433 cannot be blocked by firewall, network, or antivirus program.Please cite: http://tinyurl.com/oj7p84a
Features
- Classifies nsSNPs by pathogenicity with sensitivity of 96.3%, specificity of 79.3%, and balanced accuracy of 87.8%
- Can be used clinically to determine risk for diseases caused by nsSNPs
- Predicts nsSNP phenotype correctly 96% of time when data is available
- Provides useful annotations including UCSC Genome Browser annotations, homologous nucleotide and protein multiple sequence alignments, external nsSNP and gene information (ex. publications)
- Connected to constantly updating SQL Server which allows for fast data retrieval and up to date information
- Allows for versatile nsSNP input: amino acid location with change, dbSNP accession number, nucleotide location with change, nucleotide flanking sequence with change
- Settings allow for control of phenotype prediction, pathogenicity prediction, paralogous genes, and SQL server data retrieval
- Supports batch files
Audience
Healthcare Industry, Science/Research
User interface
Java Swing
Programming Language
Java
This is an application that can also be fetched from https://sourceforge.net/projects/gespa/. It has been hosted in OnWorks in order to be run online in an easiest way from one of our free Operative Systems.